HypoMap-an atlas of the mouse and human hypothalamus
Obesity is a growing public health problem. While our changing lifestyle and environment have undoubtedly driven this increase, there is a powerful genetic component that underlies the large variation in human body size in this modern environment. Genetic studies point to the brain, in particular a region called the hypothalamus, as having a crucial role in the regulation of appetite and body-weight. The hypothalamus can be considered the ‘fuel-sensor’ of the brain, and as a result, there have been concerted efforts within the field to understand more about this region. One of the approaches has been to try and uncover the identity of all the different cell-types within the mouse hypothalamus using single-cell sequencing approaches. Many groups, including our own (Yeo) have done this with different regions of the hypothalamus and from mice under different conditions (eg fasting or on a high-fat diet). However despite huge progress in the field, and availability of datasets in the public domain, a unified catalogue and molecular characterisation of the heterogeneous cell types and, specifically, neuronal subtypes in this brain region are still lacking.
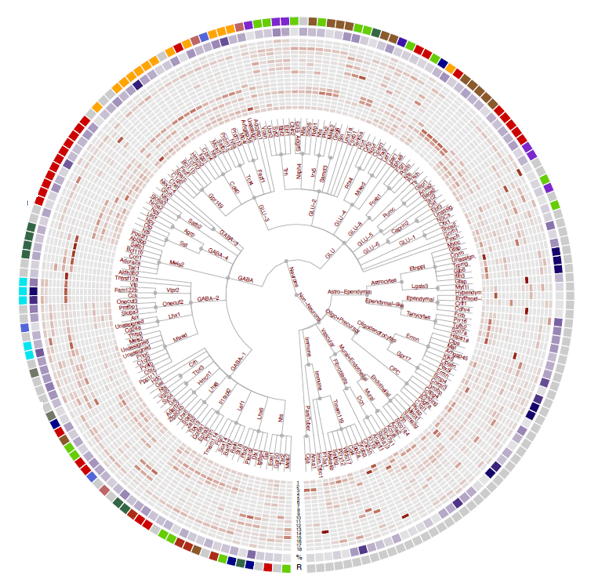
In a collaboration with colleagues at the Max Planck Institute for Metabolism Research, and published in Nature Metabolism, we have merged 18 large mouse single-cell datasets into an integrated reference atlas of the mouse hypothalamus we call ‘HypoMap’. This represents the most comprehensive database of its kind. It can serve as an important platform to unravel the functional organisation of hypothalamic circuits, and to identify novel druggable targets for treating metabolic disorders. This resource is open-access and available for anyone to use.
In a further development, we have combined single-nucleus sequencing of 433,369 human hypothalamic cells with spatial transcriptomics, generating a comprehensive spatio-cellular transcriptional map of the human hypothalamus (published in Nature).
Access the Mouse HypoMap via Cellxgene Discover (opens a new window)
Access the Human HYPOMAP (snRNASeq) and Human HYPOMAP (spatial transcriptomics) on Cellxgene
Cellxgene is a user-friendly interface that enables exploration and interrogation of single-cell data via basic point and click. It does not require any advanced bioinformatics skills (no command lines).
Tutorials on using Cellxgene
Seurat R data objects from Cambridge University’s Apollo repository: